Mutation¶
Polynomial Mutation (‘real_pm’, ‘int_pm’)¶
Details about the mutation can be found in [35]. This mutation follows the same probability distribution as the simulated binary crossover.
[1]:
from pymoo.interface import mutation
from pymoo.factory import get_mutation
import numpy as np
import matplotlib.pyplot as plt
def show(eta_mut):
a = np.full((5000, 1), 0.5)
off = mutation(get_mutation("real_pm", eta=eta_mut, prob=1.0), a)
plt.hist(off, range=(0,1), bins=200, density=True, color="red")
plt.show()
show(30)
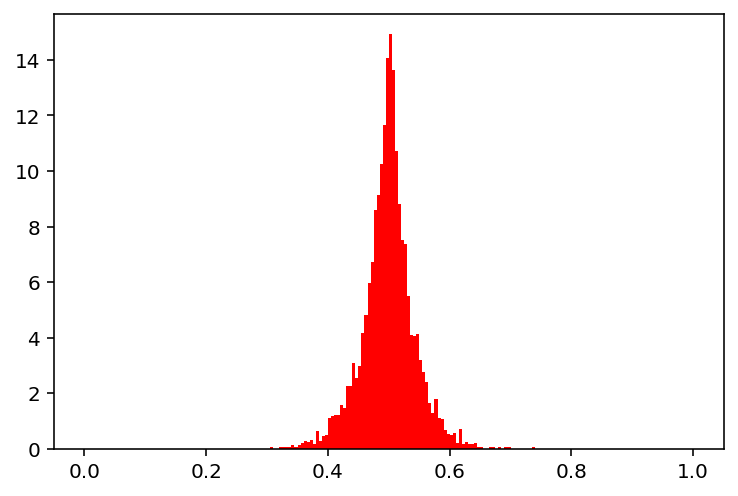
[2]:
show(10)
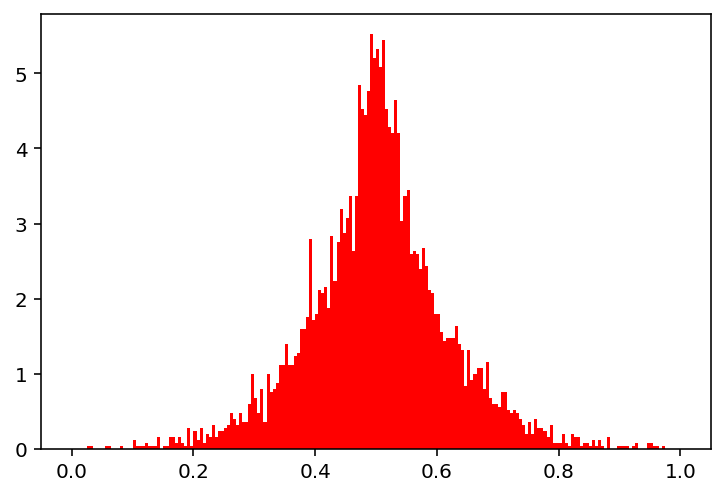
Basically, the same can be applied to discrete variables as well:
[3]:
def show(eta_mut):
a = np.full((10000, 1), 0)
off = mutation(get_mutation("int_pm", eta=eta_mut, prob=1.0), a, xl=-20, xu=20)
plt.hist(off, range=(-20, 20), bins=40, density=True, color="red")
plt.show()
show(30)
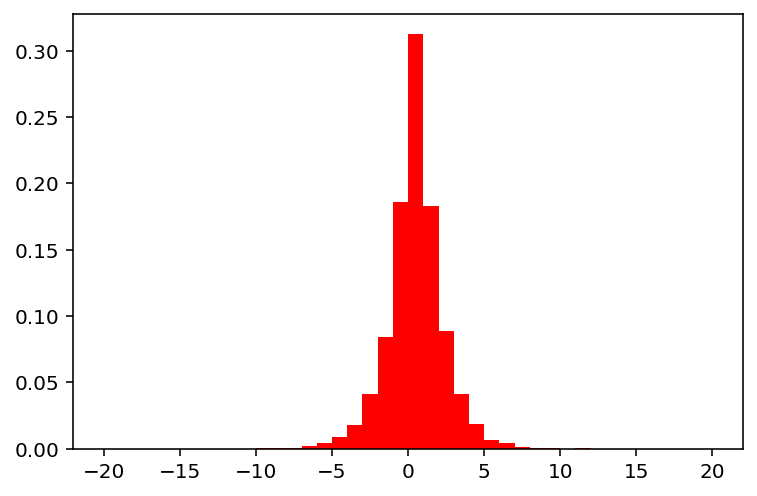
Bitflip Mutation (‘bin_bitflip’)¶
The bitlip mutation randomly flips a bit.
[4]:
def show(M):
plt.figure(figsize=(4,4))
plt.imshow(M, cmap='Greys', interpolation='nearest')
plt.show()
a = np.full((100,100), False)
mut = mutation(get_mutation("bin_bitflip", prob=0.1), a)
show(a != mut)

API¶
-
pymoo.factory.get_mutation(name, kwargs) A convenience method to get a mutation object just by providing a string.
- Parameters
- name{ ‘none’, ‘real_pm’, ‘int_pm’, ‘bin_bitflip’, ‘perm_inv’ }
Name of the mutation.
- kwargsdict
Dictionary that should be used to call the method mapped to the mutation factory function.
- Returns
- class
Mutation An mutation object based on the string. None if the mutation was not found.
- class
-
pymoo.core.mutation.Mutation() → None
