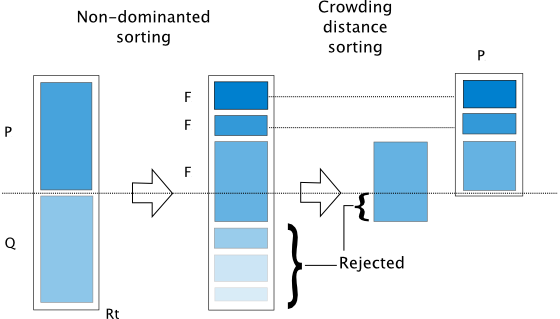
NSGA-II: Non-dominated Sorting Genetic Algorithm¶
The algorithm is implemented based on [4]. The algorithm follows the general outline of a genetic algorithm with a modified mating and survival selection. In NSGA-II, first, individuals are selected frontwise. By doing so, there will be the situation where a front needs to be split because not all individuals are allowed to survive. In this splitting front, solutions are selected based on crowding distance.
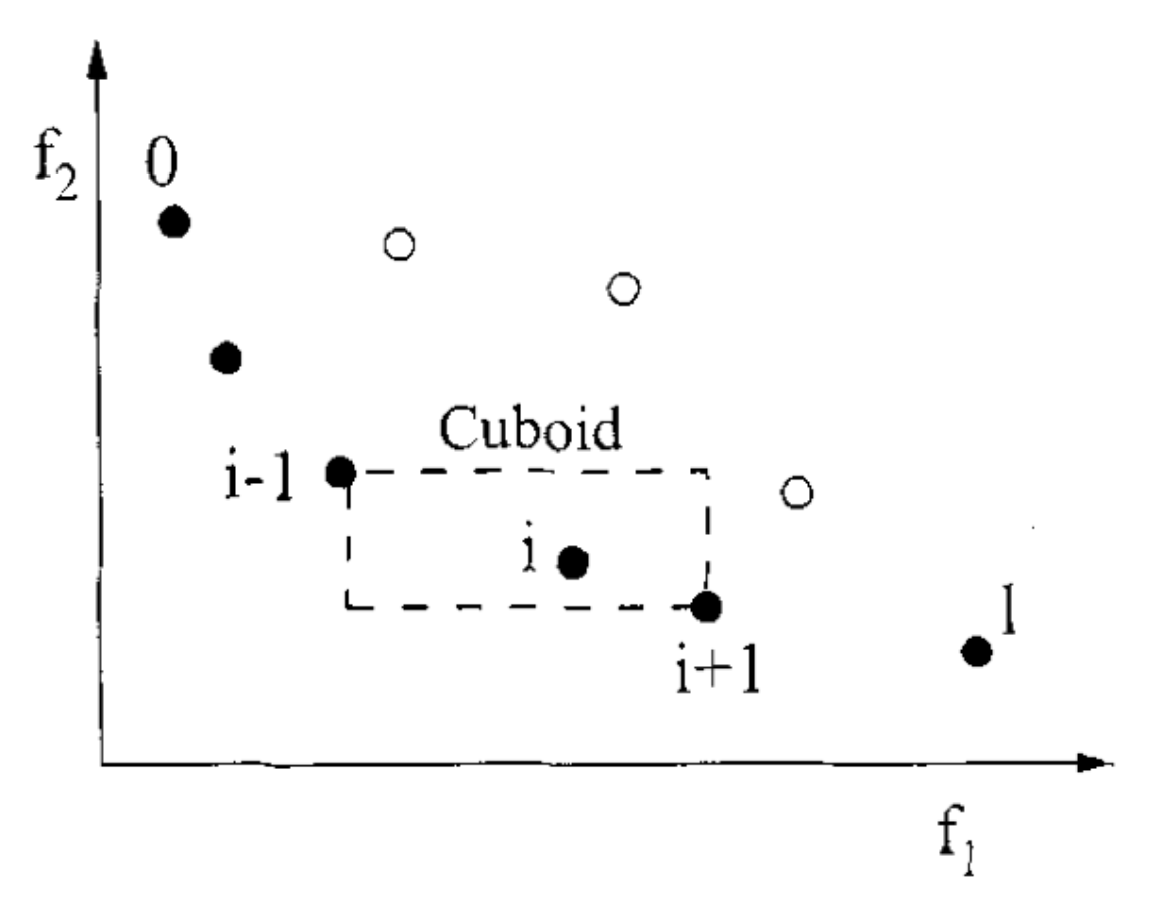
The crowding distance is the Manhatten Distance in the objective space. However, the extreme points are desired to be kept every generation and, therefore, get assigned a crowding distance of infinity.
Furthermore, to increase some selection pressure, NSGA-II uses a binary tournament mating selection. Each individual is first compared by rank and then crowding distance. There is also a variant in the original C code where instead of using the rank, the domination criterium between two solutions is used.
Example¶
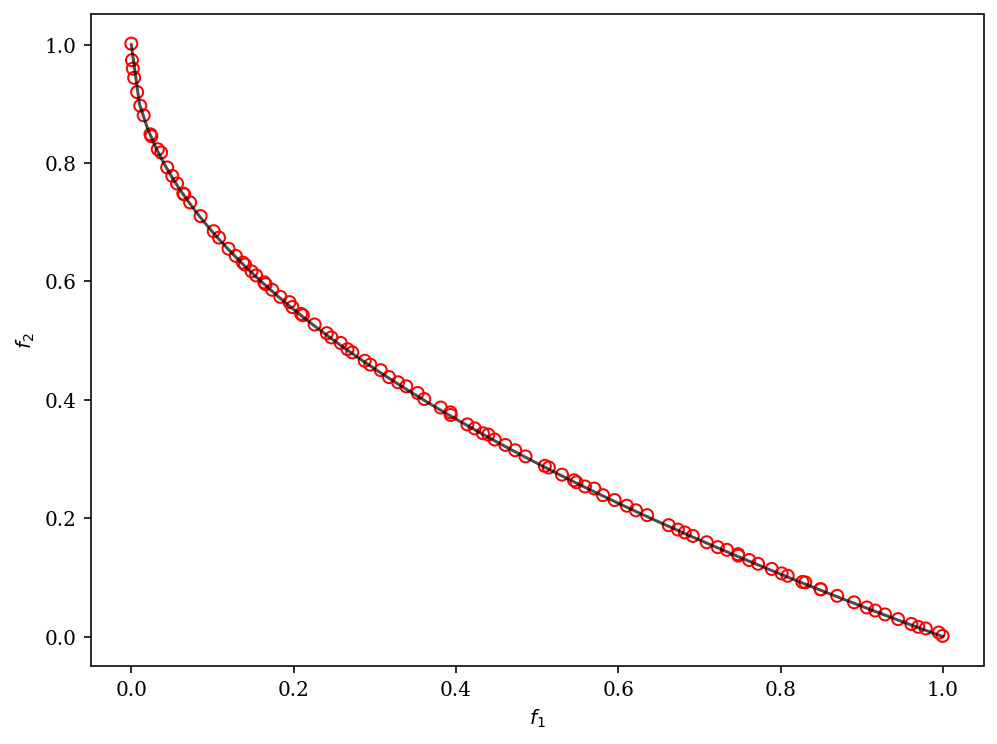
[1]:
from pymoo.algorithms.moo.nsga2 import NSGA2
from pymoo.factory import get_problem
from pymoo.optimize import minimize
from pymoo.visualization.scatter import Scatter
problem = get_problem("zdt1")
algorithm = NSGA2(pop_size=100)
res = minimize(problem,
algorithm,
('n_gen', 200),
seed=1,
verbose=False)
plot = Scatter()
plot.add(problem.pareto_front(), plot_type="line", color="black", alpha=0.7)
plot.add(res.F, facecolor="none", edgecolor="red")
plot.show()
[1]:
<pymoo.visualization.scatter.Scatter at 0x7fd56fd076a0>
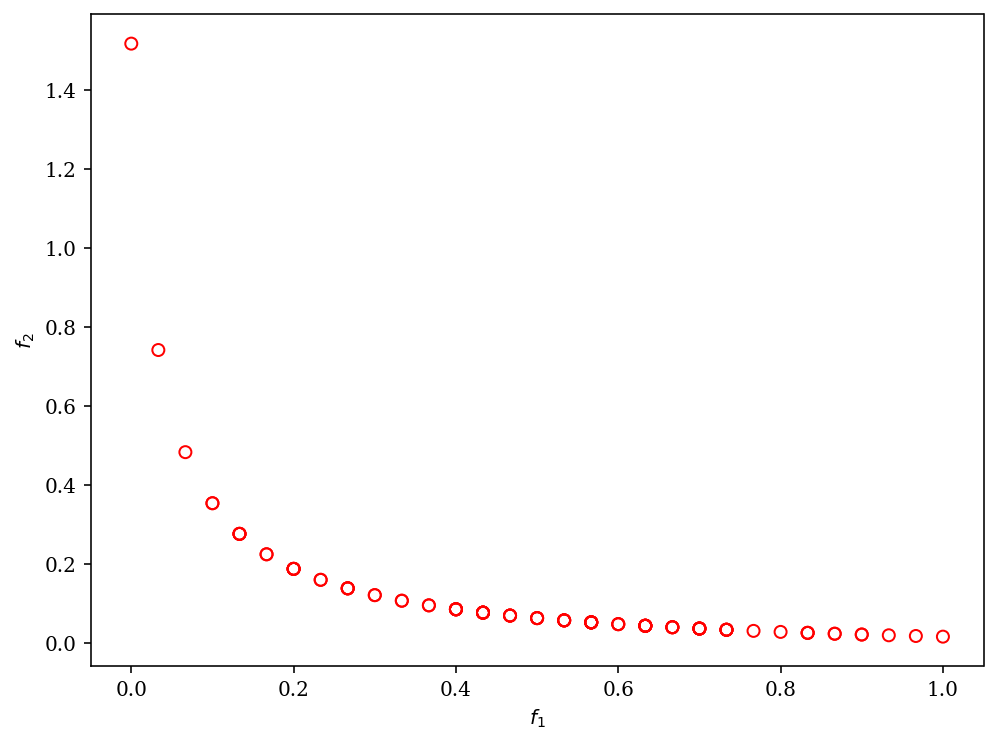
Moreover, we can customize NSGA-II to solve a problem with binary decision variables, for example, ZDT5.
[2]:
from pymoo.algorithms.moo.nsga2 import NSGA2
from pymoo.factory import get_problem, get_sampling, get_crossover, get_mutation
from pymoo.optimize import minimize
from pymoo.visualization.scatter import Scatter
problem = get_problem("zdt5")
algorithm = NSGA2(pop_size=100,
sampling=get_sampling("bin_random"),
crossover=get_crossover("bin_two_point"),
mutation=get_mutation("bin_bitflip"),
eliminate_duplicates=True)
res = minimize(problem,
algorithm,
('n_gen', 500),
seed=1,
verbose=False)
plot = Scatter()
plot.add(res.F, facecolor="none", edgecolor="red")
plot.show()
[2]:
<pymoo.visualization.scatter.Scatter at 0x7fd56fce36a0>
API¶
-
class
pymoo.algorithms.moo.nsga2.NSGA2(self, pop_size=100, sampling=FloatRandomSampling(), selection=TournamentSelection(func_comp=binary_tournament), crossover=SimulatedBinaryCrossover(eta=15, prob=0.9), mutation=PolynomialMutation(prob=None, eta=20), survival=RankAndCrowdingSurvival(), eliminate_duplicates=True, n_offsprings=None, display=MultiObjectiveDisplay(), **kwargs) - Parameters
- pop_sizeint
The population sized used by the algorithm.
- sampling
Sampling,Population,numpy.array The sampling process defines the initial set of solutions which are the starting point of the optimization algorithm. Here, you have three different options by passing
(i) A
Samplingimplementation which is an implementation of a random sampling method.(ii) A
Populationobject containing the variables to be evaluated initially OR already evaluated solutions (F needs to be set in this case).(iii) Pass a two dimensional
numpy.arraywith (n_individuals, n_var) which contains the variable space values for each individual.- selection
Selection This object defines the mating selection to be used. In an evolutionary algorithm each generation parents need to be selected to produce new offsprings using different recombination and mutation operators. Different strategies for selecting parents are possible e.g. selecting them just randomly, only in the neighborhood, using a tournament selection to introduce some selection pressure, …
- crossover
Crossover The crossover has the purpose of create offsprings during the evolution. After the mating selection the parents are passed to the crossover operator which will dependent on the implementation create a different number of offsprings.
- mutation
Mutation Some genetic algorithms rely only on the mutation operation. However, it has shown that increases the performance to perform a mutation after creating the offsprings through crossover as well. Usually the mutation operator needs to be initialized with a probability to be executed. Having a high probability of mutation will most of the time increase the diversity in the population.
- eliminate_duplicatesbool
The genetic algorithm implementation has a built in feature that eliminates duplicates after merging the parent and the offspring population. If there are duplicates with respect to the current population or in the offsprings itself they are removed and the mating process is repeated to fill up the offsprings until the desired number of unique offsprings is met.
- n_offspringsint (default: None)
Number of offspring that are created through mating. By default n_offsprings=None which sets the number of offsprings equal to the population size. By setting n_offsprings=1 a, so called, steady-state version of an algorithm can be achieved.
